Detailed variantCaller plugin report
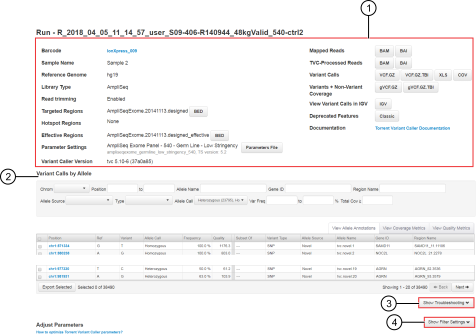
The detailed variantCaller plugin report contains variantCaller run information, results, and the associated files for download that are specific for an individual barcode or sample.
To access the report, click the sample name link in the Sample column in a report from a sequencing run.
-
Review run information for a specific barcode or sample and download the associated files.
-
View variants called and their associated allele annotation information, coverage metrics, and quality metrics. For more information, see Variant Calls by Allele table.
-
Export the variant data files for troubleshooting. For more information, see Export files for troubleshooting.
-
Adjust variantCaller plugin filter settings that were used for the specific barcode or sample, then save the adjusted parameters to a new configuration. For more information, see Save adjusted parameters to a variantCaller plugin configuration.
The following table lists and describes the download options for an individual barcode or the sample used in the run. The available options in the table depend on the run type.
