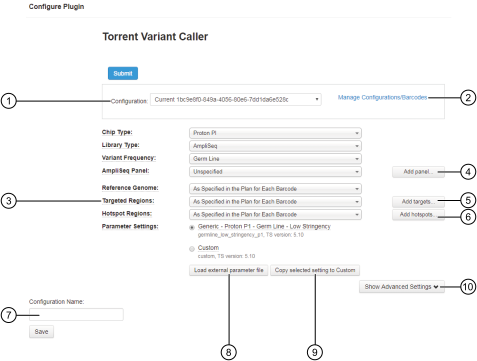
variantCaller plugin configuration
Use the Torrent Variant Caller screen to configure the variantCaller plugin. For information about how to get to this screen, see Configure the variantCaller plugin to run as part of a Planned Run
|
Setting |
Details |
|---|---|
|
Configuration [1] |
A reusable predefined or a custom configuration for the variantCaller plugin that includes settings for a reference genome, targeted regions, hotspots, and parameter settings. For more information, see Create a custom configuration for the variantCaller plugin. |
|
Manage Configurations/Barcodes 1 |
Manage and apply configurations for barcodes when you run the variantCaller plugin manually. For more information, see Apply configuration settings to specific barcodes. (Optional) To run the variantCaller plugin manually for a limited number of barcodes, select Skip this barcode for the barcodes that you do not want to include in the plugin run. For more information, see Apply configuration settings to specific barcodes. |
|
Chip Type 1 |
The chip type that is used in the sequencing run. If you change this option, it affects only the fields that are selected by default in the variantCaller Configuration screen. It does not affect the sequencing run. |
|
Library Type 1 |
Library type options include: When Library Type is set to AmpliSeq, read trimming is automatically applied to remove the primers from reads. |
|
Variant Frequency 1 |
Variant frequency options include: |
|
AmpliSeq Panel 1 |
Panels ordered from AmpliSeq.com have predefined variantCaller plugin parameter settings. For details, see Import panel files and parameters. When an Ion AmpliSeq™ panel is selected, the plugin configuration screen automatically selects target regions, hotspots, and parameter settings files that are for use with the panel. |
|
Add panel 1 |
Upload a panel. |
|
Reference Genome 1 |
The reference genome that is for use with variant calling. To configure barcodes in the run to use the same genome reference that was used for the current run report, select the As Specified in the Plan for Each Barcode option. If the selected reference genome differs from the reference genome that is included in the Planned Run, the software must realign the data, which requires more time for the plugin run. |
|
Target Regions are the regions of interest for which you want to call variants. If a target regions file is not provided, the variantCaller plugin analyzes every position of the reference genome, which typically takes longer. To configure barcodes to use the same target regions file that is included in the current Planned Run, select the As Specified in the Plan for Each Barcode option. Before a targeted regions file can be selected, it must be uploaded from in Torrent Suite™ Software and associated with a specific reference genome. For more information, see Upload a target regions file. |
|
|
Upload a target regions file. |
|
|
Hotspots files are BED or VCF files that define variant alleles of interest. Hotspots files instruct the variantCaller plugin to include these variant alleles in its output files, including evidence for a variant and the filtering thresholds that disqualified a variant candidate. A hotspots file affects only the variantCaller plugin, not other parts of the analysis pipeline. If you do not specify a hotspots file, the software reports only the de novo variants that show as present. In contrast, if a hotspots file is used, the variant calls and the filtering metrics for each hotspot allele are reported in the output VCF file, including data for absent or NOCALL variants. To configure hotspots files to use the same hotspots file that is included in the current Planned Run, select the As Specified in the Plan for Each Barcode option. Before a hotspots file can be selected, it must be uploaded from in Torrent Suite™ Software and associated with a specific reference genome. For more information, see Upload a hotspots file. IMPORTANT! A carefully designed hotspot file is recommended to optimize the overall performance of variant calling. For help with designing a hotspots file, contact your local Field Service Engineer. |
|
|
Upload a hotspots file. |
|
|
Option to use parameter settings that are predefined in the variantCaller plugin, or to use Custom parameter settings. For Ion AmpliSeq™ experiments, panel templates from AmpliSeq.com can contain parameter settings that are optimized for the variantCaller plugin and are available for use during variantCaller plugin configuration.
|
|
|
Imports a file that contains parameter settings. |
|
|
Creates a custom parameter setting that is based on a generic parameter setting. For more information, see Create and use a custom parameters setting. |
|
|
Configuration Name 1 |
Names and saves a custom configuration when changes are made to the predefined settings. |
|
Configure advanced parameter settings. For more information, see variantCaller plugin advanced parameters. Hover your mouse over the field to see tooltips with descriptions of the advanced settings. |