Review ampliSeqRNA plugin results
The ampliSeqRNA plugin generates an initial summary report that lists the samples, the number of mapped reads, the percent of valid reads, and the percent of targets detected. A series of log2 reads-per-million (RPM) pair correlation plots are included for rapid correlation analysis. Microsoft™ Excel™-compatible reports are also generated, including differential expression tables. Additional details about read coverage are also provided on a per-barcode basis, along with a list of gene annotations for each sequenced region.
After the sequencing run completes, review the plugin results in the report summary.
- In the Data tab, click Completed Runs & Reports.
- In the list of runs, locate the run of interest, then click the link in the Report Name column.
-
In the left navigation menu, click ampliSeqRNA to view the plugin results.
-
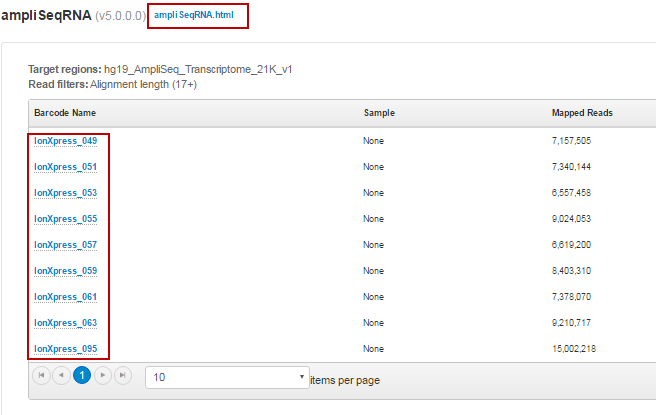
Click the ampliSeqRNA.html link to open the ampliSeqRNA Report – Barcode Summary for all barcodes.
-
In the barcode table, click individual barcode names to see the results for an individual barcode.
-
Click the Distribution Plots, Correlation Heatmap, Correlation Plot, and Gene Heatmap tabs to review the data graphically.
-
Click the links at the bottom of the report to download associated report files.
-