Review immuneResponseRNA plugin results
- In the Data tab, click Completed Runs & Reports.
- In the list of runs, find the run of interest, then click the link in the Report Name column.
- In the left navigation menu, click immuneResponseRNA to view the plugin summary.
-
In the immuneResponseRNA section, click the immuneResponseRNA.html link to open the immuneResponseRNA Report for all barcodes.
BAM files load quickly, and you may see these files first in the list of links. The other file formats take longer to download, so you may have to wait for the links to the VCF, XLS, and FASTQ formats to appear.
-
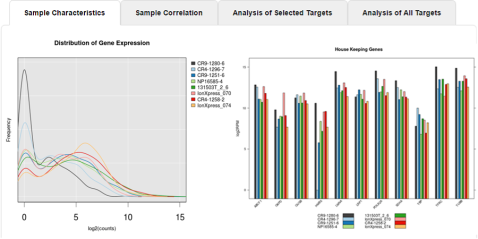
In the Analysis Summary window, review your Mapped Reads, Valid Reads, and the Targets that are detected by barcode.
Column
Description
The barcode used for the sample.
The sample name as it was entered in the sequencing Run Plan.
The number of reads that map to the reference sequences.
The percentage of mapped reads ≥50% amplicon length.
The number of targets/genes with at least 1 read.
The number of targets/genes with at least 2 reads.
The number of targets/genes with at least 10 reads.
- Click an individual barcode name to view the results for that barcode.
- Scroll down, then click the Sample Characteristics, Sample Correlation, Analysis of Selected Targets (available only if a Genes of interest subset BED file was selected), or Analysis of All Targets tabs to review the data in graphic format.