RNASeqAnalysis plugin
The RNASeqAnalysis plugin is an RNA Transcript Alignment and Analysis tool for use with reference genomes hg19 and mm10.
To use the hg19 or mm10 genomes with this plugin, the reference genomes must first be imported from the preloaded references screen. Also, annotation files for the human and mouse references are available for import. For more information on downloading references and annotation files, see Import a preloaded reference sequence file.
|
Annotation file name |
Description |
|---|---|
|
Human genome annotation (hg19) |
|
|
Human auxiliary small RNA sequences (cDNA) |
|
|
Mouse genome annotation (mm10) |
|
|
Mouse auxiliary small RNA sequences (cDNA) |
|
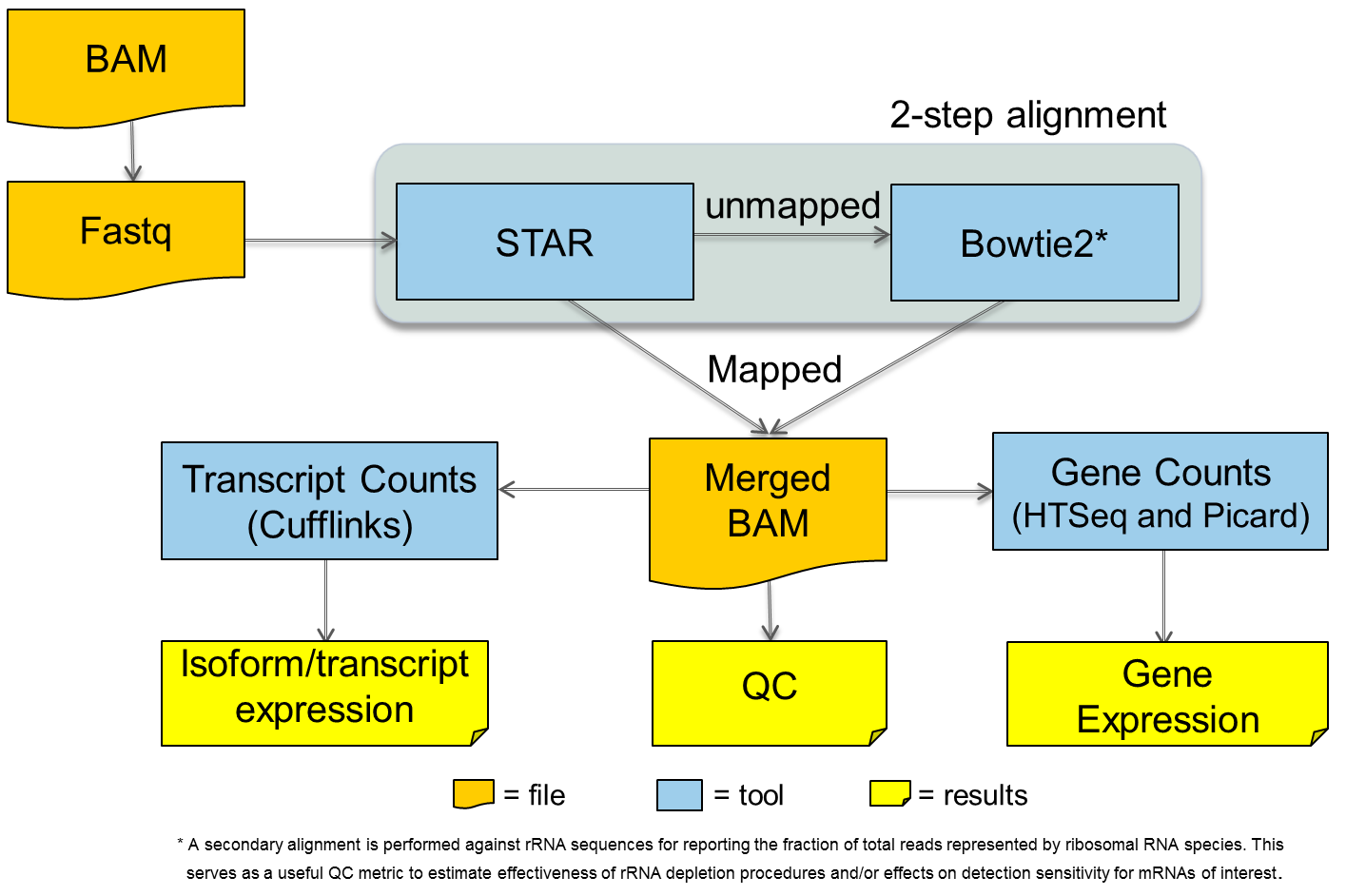
Use this plugin to analyze cDNA reads, as produced by RNA-Seq. Reads are aligned to the reference genome using STAR and bowtie2 aligners to find full and partial mappings. The alignments are analyzed by HTSeq and Picard tools to collect assigned read counts and cufflinks to extract gene isoform representation. For barcoded data, comparative representation plots across barcodes are created in addition to individual reports for each barcode. All alignment, detail, and summary report files are available for download.