Read filtering and trimming
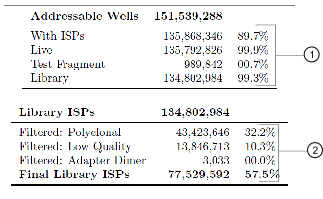
Empty and loaded wells are separated by the differences in buffering and signal over the chip during the nucleotide key flows (flows 1–8). Wells that are loaded with ISPs and associated polymerase have greater buffering capacity and higher signal than empty wells, and Torrent Suite™ Software uses these differences to identify and classify loaded versus empty wells.
After well classification, the software further processes the identified test fragments and library reads, including read filtering and trimming. This processing affects the total number of library reads and bases. You can see both well classification and library read filtering results that are displayed in the run report. Read trimming operations trim bases off the read, thereby making reads shorter, while read filtering operations completely remove them from the output BAM files. By default, reads that have a trimmed read length of less than 25 bases are being filtered. The different categories of filtered reads shown in the run report are in the table below.
|
Filter |
Description |
|---|---|
|
Polyclonal |
Filters reads from ISPs with >1 unique library template population. Occasionally, low or unexpected signal ISPs can also get caught in this filter. |
|
Low Quality |
Filters reads with unrecognizable key signal, low signal quality, and reads trimmed to <25 bases. |
|
Primer Dimer |
Filters reads where no or only a very short sequencing insert is present. Reads that, after P1 adapter trimming, have a trimmed length of <25 bases are considered primer dimers. |
You can see both well classification and library read filtering results that are displayed in the run report.
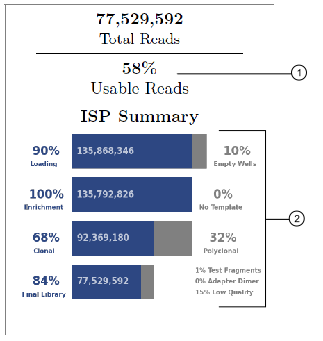
The ISP summary panel also presents the well classification and library ISP summary table using slightly different calculations.
The usable reads percentage is calculated by
Each ISP summary percentage is calculated by dividing the current value by the previous values. For example,