Find false negatives with an alignment viewer
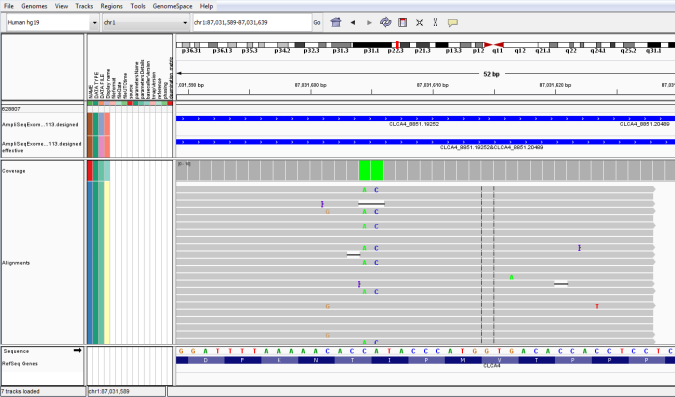
When an expected variant is not called by the variantCaller plugin, an alignment viewer, such as Integrative Genomics Viewer (IGV) or Ion Reporter™ Genomic Viewer (IRGV) can help you confirm the absence of the variant in the sample, or help you understand how to adjust the plugin parameters to enable the plugin to call the variant. Using the IGV or IRGV can reveal problems such as mismapping or low coverage. In particular, an alignment viewer lets you visually inspect the coverage of the region where the variant is expected, and focus attention on the depth of coverage and the quality of the bases covering the position of the variant. Low coverage or low base quality can explain a no-call. A genomic viewer can also reveal that variant is slightly misplaced (especially for INDELs) and therefore not called.
- On the detailed variantCaller plugin summary report screen, click IGV to open the viewer.
- In the viewer, select the chromosome where the variant of interest is located, scroll to the chromosomal position of the variant, then zoom in until you can read the nucleotide sequence surrounding the variant allele.
- Hover over a variant read to view read analytics.
- Adjust plugin parameters.
- Rerun the variantCaller plugin.